Abstract
BACKGROUND: Autophagy plays an important role in multiple cell processes including elimination of misfolded proteins and damaged organelles, clearance of intracellular microbes and regulation of innate immunity. Autophagic activity is significantly up regulated in diseases characterized by proteins aggregation such as Alzheimer's, Huntington's, and Parkinson's diseases. Sickle cell anemia (SCA) is associated with E6V mutation in beta-globin gene that induces hemoglobin (Hb) polymerization under low oxygen or acidic conditions. SCA is characterized by chronic presence of low-level plasma Hb (3-10 µM). Hb is cleared from circulation by macrophage's endocytosis, resulting in degradation of Hb in lysosomes. Acidic lysosome environment may induce Hb polymerization, lysosome dysregulation and autophagy activation.
HYPOTHESIS: We hypothesize that endocytosis of HbS induces autophagy in human macrophages.
METHODS: The study was approved by Howard University review board (IRB) and all subjects provided written inform consent prior the sample collection. Whole blood samples were obtained from three SCA patients and healthy individuals. Human THP-1 promonocytic cells were differentiated into macrophages with 25 nM PMA for 72 hrs and treated with either purified HbS or HbA (5 µM, Sigma-Aldrich). Total RNA was depleted of cytoplasmic and mitochondrial ribosomal RNA, and strand-specific libraries were constructed using the Illumina ® TruSeq Stranded Total RNA Gold kit. Libraries were sequenced on an Illumina ® NextSeq 500 using 75 bp paired-end sequencing on two v2.5 150 cycle High-Output kits, generating 40-50 million paired-end reads per sample. The sequencing data were mapped to human reference genome using Dragen RNA v.3.8.4 software (Illumina). Comparisons were conducted using Dragen differential expression software v. 3.6.3 software (Illumina). Transmission electron microscopy (TEM) imaging was performed at Talos 200X transmission electron microscope (Thermo Fisher Scientific). CYTO-ID Autophagy detection kit (Enzo Life Science) was used for detection of autophagy in live cells by fluorescence microscopy, and fluorescent microplate assay.
RESULTS: In THP-1-derived macrophages treated with HbS compared to HbA, 28362 genes were detected including 322 differentially expressed genes (1.1%, 1.5-fold difference, 187 down and 135 up) at 5% FDR. In THP-1-differentiated macrophages treated with SCD RBC lysate compared to control RBC lysate, 30840 genes were detected including 256 differentially expressed genes (0.83%, 1.5-fold difference, 117 down and 139 up) at 5% FDR.
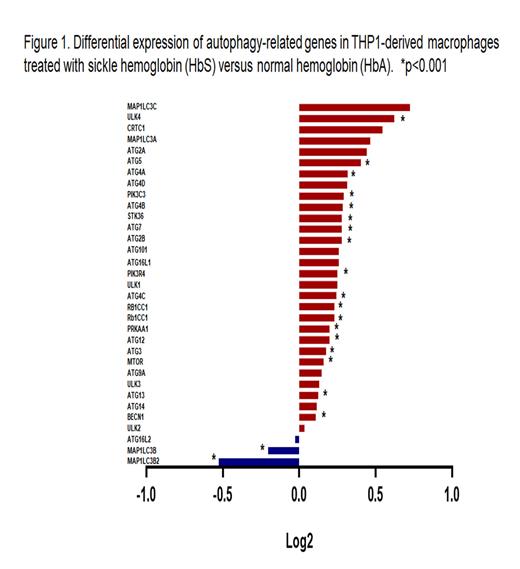
We focused our analysis on 33 autophagy-related genes. Among these 33 genes, 30 genes were up-regulated (1.2-1.7-fold) and only 3 genes were down-regulated in macrophages treated with HbS compared to HbA (Fig.1). These changes were statistically significant for 20 genes (p<0.01). No significant differences were found in expression of autophagy-related genes in macrophages treated either with HbS or SCD RBC lysate.
Autophagy activation was validated by CYTO-ID Autophagy detection kit, demonstrating significant autophagy activation in HbS treated macrophages compared to HbA treatment (1.2-fold, p=0.0019). Transmission electron microscopy demonstrated accumulation of autophagosomes in the cells treated with HbS compared to non-treated cells and cells treated with HbA.
CONCLUSION: Treatment of macrophages with sickle cell hemoglobin or lysate of red blood cells collected from SCD patients up-regulates expression of autophagy-related genes and increased autophagosome formation.
ACKNOWLEDGMENTS: We thank Drs. Castle Raley and Keith Crandall, George Washington University SPH Genomics Core, for sequencing and guidance in data analysis; Dr. Christine Brantner, GW Nanofabrication and Imaging Center, for TEM imaging; and students Kavita Pandya, Gretchen Whitney High School, Cerritos, CA, and Tharindi Jayatilake, Montgomery Blair High School, Silver Spring, MD, for help in analysis of autophagy-related gene expression.
No relevant conflicts of interest to declare.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal